1.What the tool can do for you?
This tool focuses on exploring correlation between probesets in gene expression data and compare with clinical data available.
Original paper: Horvath et al, BMC Bioinformatics 2008, 9:559 doi:10.1186/1471-2105-9-559 'WGCNA: an R package for weighted correlation network analysis'
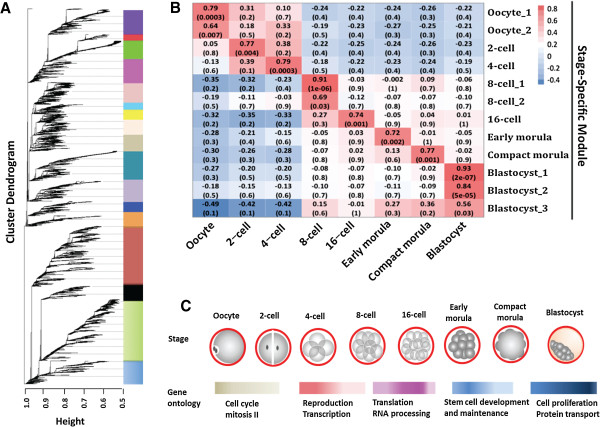
The following image is from: Jiang et al, BMC Genomics 2014 15:756 doi:10.1186/1471-2164-15-756, 'Transcriptional profiles of bovine in vivo pre-implantation development'
5.Acknowledgements
We would like to acknowledge the authors of the following R packages (WGCNA, rmarkdown, plotly, shiny, stringr, DT, NetworkD3, gplots,
impute) needed for the main app, the author of the radiant R package for the inspiration for the save/load functionality and the authors
of the paper “Integrating Genetic and Network Analysis to Characterize Genes Related to Mouse Weight”, PLoS Genetics, 2006, for publishing
the data used as demo datasets.


